Hi,
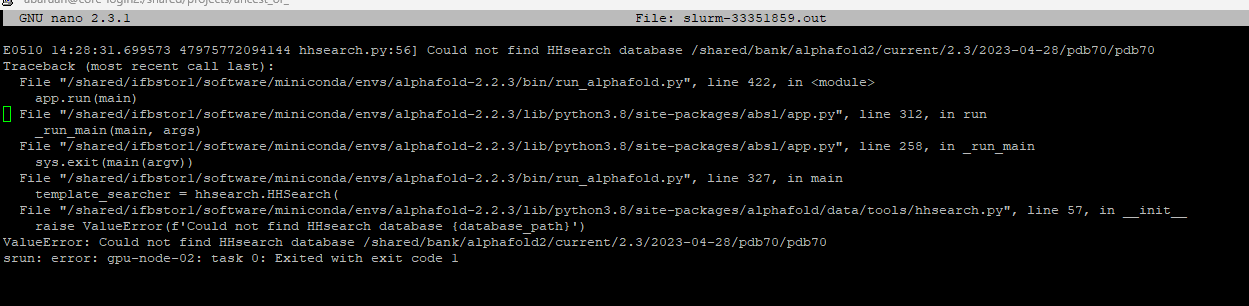
Both versions of Alphafold2 are not working in the IFB cluster and all my jobs were killed. I have attached an Error screenshot for both. can you please help in this matter
Thanking you
Here is my script for Alphafold 2.1.1
#!/bin/bash
#SBATCH --partition=gpu
#SBATCH --gres=gpu:7g.40gb:1
#SBATCH --cpus-per-task=10
#SBATCH --mem=50G
#SBATCH --account=ancest_or_
#SBATCH --time 36:00:00
module load alphafold/2.1.1
mkdir -p /tmp/$USER_alphafold
srun run_alphafold.sh --fasta_paths=/shared/projects/ancest_or_/SfruGR274.fa,SfruGR213.fa,SfruGR210.fa,SfruGR271.fa,SfruGR230.fa \
--output_dir=/shared/projects/ancest_or_ \
--model_preset=monomer \
--db_preset=full_dbs \
--data_dir=/shared/bank/alphafold2/current/2.3/2023-04-28 \
--uniref90_database_path=/shared/bank/alphafold2/current/2.3/2023-04-28/uniref90/uniref90.fasta \
--mgnify_database_path=/shared/bank/alphafold2/current/2.3/2023-04-28/mgnify/mgy_clusters_2018_12.fa \
--pdb70_database_path=/shared/bank/alphafold2/current/2.3/2023-04-28/pdb70/pdb70 \
--template_mmcif_dir=/shared/bank/alphafold2/current/2.3/2023-04-28/pdb_mmcif/mmcif_files \
--max_template_date=2020-05-14 \
--obsolete_pdbs_path=/shared/bank/alphafold2/current/2.3/2023-04-28/pdb_mmcif/obsolete.dat \
--bfd_database_path=/shared/bank/alphafold2/current/2.3/2023-04-28/bfd/bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt \
--uniclust30_database_path=/shared/bank/alphafold2/2022-09-21/uniclust30/uniclust30_2018_08/uniclust30_2018_08
Here is my script for Alphafold 2.2.3
#!/bin/bash
#SBATCH --partition=gpu
#SBATCH --gres=gpu:3g.20gb:1
#SBATCH --cpus-per-task=10
#SBATCH --mem=50G
#SBATCH --account=ancest_or_
#SBATCH --time 36:00:00
module load alphafold/2.2.3
mkdir -p /tmp/$USER_alphafold
srun run_alphafold.sh --use_gpu_relax=True --fasta_paths=/shared/projects/ancest_or_/SfruGR180.fa,SfruGR269.fa,SfruGR186.fa,SfruGR187.fa \
--output_dir=/shared/projects/ancest_or_ \
--model_preset=monomer \
--db_preset=full_dbs \
--data_dir=/shared/bank/alphafold2/current/2.3/2023-04-28 \
--uniref90_database_path=/shared/bank/alphafold2/current/2.3/2023-04-28/uniref90/uniref90.fasta \
--mgnify_database_path=/shared/bank/alphafold2/current/2.3/2023-04-28/mgnify/mgy_clusters_2018_12.fa \
--pdb70_database_path=/shared/bank/alphafold2/current/2.3/2023-04-28/pdb70/pdb70 \
--template_mmcif_dir=/shared/bank/alphafold2/current/2.3/2023-04-28/pdb_mmcif/mmcif_files \
--max_template_date=2020-05-14 \
--obsolete_pdbs_path=/shared/bank/alphafold2/current/2.3/2023-04-28/pdb_mmcif/obsolete.dat \
--bfd_database_path=/shared/bank/alphafold2/current/2.3/2023-04-28/bfd/bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt \
--uniclust30_database_path=/shared/bank/alphafold2/2022-09-21/uniclust30/uniclust30_2018_08/uniclust30_2018_08
Hello @Abhinob_Baruah,
/shared/bank/alphafold2/current/2.3/2023-04-28/pdb70/pdb70 does not exist, but /shared/bank/alphafold2/current/2.3/pdb70 does.
Can you try with this database path ? (or any valid database path in /shared/bank/alphafold2 if you need to use a different version of the pdb)
1 « J'aime »
n'est ce pas plutot un problème de droit de lecture sur la bank:
cp: impossible d'ouvrir « /shared/bank/alphafold2/2023-04-28/uniref30/UniRef30_2021_03_a3m.ffdata » en lecture: Permission non accordée
cp: impossible d'ouvrir « /shared/bank/alphafold2/2023-04-28/uniref30/UniRef30_2021_03_hhm.ffindex » en lecture: Permission non accordée
cp: impossible d'ouvrir « /shared/bank/alphafold2/2023-04-28/uniref30/UniRef30_2021_03_a3m.ffindex » en lecture: Permission non accordée
cp: impossible d'ouvrir « /shared/bank/alphafold2/2023-04-28/uniref30/UniRef30_2021_03_cs219.ffindex » en lecture: Permission non accordée
cp: impossible d'ouvrir « /shared/bank/alphafold2/2023-04-28/uniref30/UniRef30_2021_03.md5sums » en lecture: Permission non accordée
cp: impossible d'ouvrir « /shared/bank/alphafold2/2023-04-28/uniref30/UniRef30_2021_03_cs219.ffdata » en lecture: Permission non accordée
cp: impossible d'ouvrir « /shared/bank/alphafold2/2023-04-28/uniref30/UniRef30_2021_03_hhm.ffdata » en lecture: Permission non accordée
j'ai la meme erreur lorsque je fais tourner alphafold
Hi,
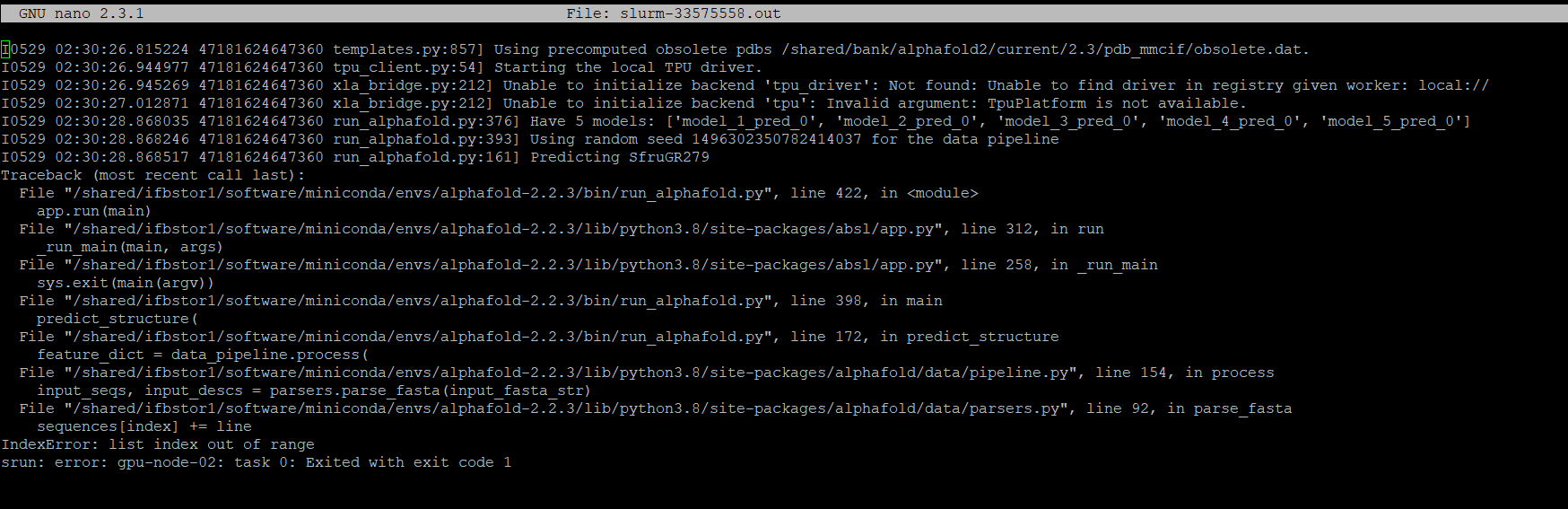
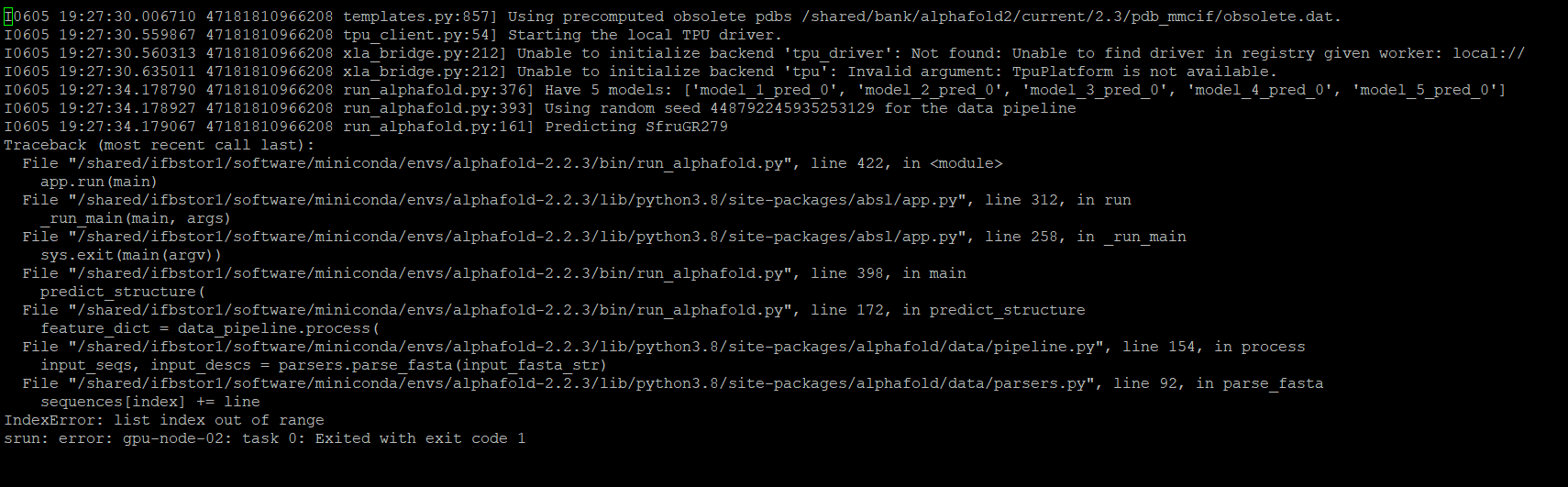
I have enountreed a Error again with the current scirpt,my job was killed. Please can you kindly let me know what to do? I need to complete my experiments in time for my M2 master thesis Deadline. I hope you will help me in this matter. I really appreciate it, I hope will hear from your team soon
Thank you
Here is my script,
#!/bin/bash
#SBATCH --partition=gpu
#SBATCH --gres=gpu:3g.20gb:1
#SBATCH --cpus-per-task=10
#SBATCH --mem=50G
#SBATCH --account=ancest_or_
#SBATCH --time 36:00:00
module load alphafold/2.2.3
mkdir -p /tmp/$USER_alphafold
srun run_alphafold.sh --use_gpu_relax=True --fasta_paths=/shared/projects/ancest_or_/SfruGR279.fa,SfruGR15.fa,OfurGR47.fa,OfurOR34.fa
--output_dir=/shared/projects/ancest_or_
--model_preset=monomer
--db_preset=full_dbs
--data_dir=/shared/bank/alphafold2/current/2.3
--uniref90_database_path=/shared/bank/alphafold2/current/2.3/uniref90/uniref90.fasta
--mgnify_database_path=/shared/bank/alphafold2/current/2.3/mgnify/mgy_clusters_2018_12.fa
--pdb70_database_path=/shared/bank/alphafold2/current/2.3/pdb70/pdb70
--template_mmcif_dir=/shared/bank/alphafold2/current/2.3/pdb_mmcif/mmcif_files
--max_template_date=2020-05-14
--obsolete_pdbs_path=/shared/bank/alphafold2/current/2.3/pdb_mmcif/obsolete.dat
--bfd_database_path=/shared/bank/alphafold2/current/2.3/bfd/bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt
--uniclust30_database_path=/shared/bank/alphafold2/2022-09-21/uniclust30/uniclust30_2018_08/uniclust30_2018_08
Hi @Abhinob_Baruah,
It's not an issue with Alphafold, the command looks like it failed because of the argument --fasta_paths=/shared/projects/ancest_or_/SfruGR279.fa,SfruGR15.fa,OfurGR47.fa,OfurOR34.fa you provided.
Can you try with this: --fasta_paths=/shared/projects/ancest_or_/SfruGR279.fa,/shared/projects/ancest_or_/SfruGR15.fa,/shared/projects/ancest_or_/OfurGR47.fa,/shared/projects/ancest_or_/OfurOR34.fa instead ? (what I assume are the absolute paths to your fasta sequences)
Yes, you are right that is the path to my fasta file of my gene models. But, it's very strange that even after changing the script my Job was pending for almost 2 days, which is not usual, and neither did it take so long. Please, can you let me know what is the problem here? I really appreciate it.
Thanks
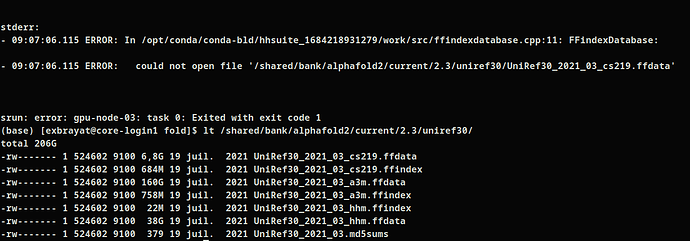
Hi , I have a problem that seems similar to me on alphafold 2.3.2, it seems the permission are not set correctly, did not have read permission for "/shared/bank/alphafold2/current/2.3/uniref30/" files. Is this the issue ? How can i contact admins to change the permissions ?
Thanks for your help
Antoni
Hello,
I also had difficulties running Alphafold2 after the change in databases (I had similar access rights issues).
With a lot of trial and error of changing the databases I got the monomer mode to work, but now I have an error running the multimer mode due to the version of the multimer param files and I am not sure how to solve it.
cat slurm-33630046.out
I0602 11:33:02.082411 47616662142656 templates.py:857] Using precomputed obsolete pdbs /shared/bank/alphafold2/current/2.3/pdb_mmcif/obsolete.dat.
Traceback (most recent call last):
File "/shared/ifbstor1/software/miniconda/envs/alphafold-2.2.3/bin/run_alphafold.py", line 422, in <module>
app.run(main)
File "/shared/ifbstor1/software/miniconda/envs/alphafold-2.2.3/lib/python3.8/site-packages/absl/app.py", line 312, in run
_run_main(main, args)
File "/shared/ifbstor1/software/miniconda/envs/alphafold-2.2.3/lib/python3.8/site-packages/absl/app.py", line 258, in _run_main
sys.exit(main(argv))
File "/shared/ifbstor1/software/miniconda/envs/alphafold-2.2.3/bin/run_alphafold.py", line 370, in main
model_params = data.get_model_haiku_params(
File "/shared/ifbstor1/software/miniconda/envs/alphafold-2.2.3/lib/python3.8/site-packages/alphafold/model/data.py", line 30, in get_model_haiku_params
with open(path, 'rb') as f:
FileNotFoundError: [Errno 2] No such file or directory: '/shared/bank/alphafold2/current/2.3/params/params_model_1_multimer_v2.npz'
srun: error: gpu-node-02: task 0: Exited with exit code 1
That directory only contains v3 files:
ls /shared/bank/alphafold2/current/2.3/params/
LICENSE params_model_1.npz params_model_2_multimer_v3.npz params_model_2_ptm.npz params_model_3.npz params_model_4_multimer_v3.npz params_model_4_ptm.npz params_model_5.npz
params_model_1_multimer_v3.npz params_model_1_ptm.npz params_model_2.npz params_model_3_multimer_v3.npz params_model_3_ptm.npz params_model_4.npz params_model_5_multimer_v3.npz params_model_5_ptm.npz
Is there a way to work with them?
Here is my batch script:
#!/bin/bash
# Author : Domitille JARRIGE
# Date : 2023-06-02
#
# Batch Script to run AlphaFold2 on user protein fasta file sequences on IFB cluster.
# The user also has to define an output directory for AlphaFold2 prediction.
#___________________________________________________________________________________________________________
# USAGE : sbatch alphafold.sh -q <query protein fasta file> -o <output directory>
# -m <AlphaFold mode: one of 'monomer', 'monomer_casp14', 'monomer_ptm' or 'multimer'>
#___________________________________________________________________________________________________________
#SBATCH -p gpu
#SBATCH --gres=gpu:7g.40gb
#SBATCH --cpus-per-task=10
#SBATCH --mem=100G
#SBATCH -A metacloud
#SBATCH --time 24:00:00
path=$(pwd)
while getopts hq:o:m: flag
do
case "${flag}" in
q) query_file=${OPTARG};;
o) out_dir=${OPTARG};;
m) mode=${OPTARG};;
h) echo "USAGE: sbatch alphafold.sh -q <query protein fasta file> -o <output directory> -m <AlphaFold mode on of 'monomer', 'monomer_casp14', 'monomer_ptm' or 'multimer'>"
exit 0;;
*) echo "USAGE: sbatch alphafold.sh -q <query protein fasta file> -o <output directory> -m <AlphaFold mode on of 'monomer', 'monomer_casp14', 'monomer_ptm' or 'multimer'>"
exit 1;;
esac
done
module load alphafold/2.2.3
mkdir -p /tmp/$USER_alphafold
if [[ "${mode}" == "monomer" ]]; then
param='--pdb70_database_path=/shared/bank/alphafold2/current/2.3/pdb70/pdb70'
elif [[ "${mode}" == "multimer" ]]; then
param='--uniprot_database_path=/shared/bank/alphafold2/current/2.3/uniprot/uniprot.fasta --pdb_seqres_database_path=/shared/bank/alphafold2/current/2.3/pdb_seqres/pdb_seqres.txt'
else
param='--pdb70_database_path=/shared/bank/alphafold2/current/2.3/pdb70/pdb70'
fi
srun run_alphafold.sh --fasta_paths="${path}/${query_file}" \
--output_dir="${path}/${out_dir}" \
--model_preset="${mode}" \
--use_gpu_relax=True \
--db_preset=full_dbs \
--data_dir=/shared/bank/alphafold2/current/2.3/ \
--uniref90_database_path=/shared/bank/alphafold2/current/2.3/uniref90/uniref90.fasta \
--mgnify_database_path=/shared/bank/alphafold2/current/2.3/mgnify/mgy_clusters_2018_12.fa \
--template_mmcif_dir=/shared/bank/alphafold2/current/2.3/pdb_mmcif/mmcif_files \
--max_template_date=2020-05-14 \
--use_precomputed_msas=True \
--obsolete_pdbs_path=/shared/bank/alphafold2/current/2.3/pdb_mmcif/obsolete.dat \
--bfd_database_path=/shared/bank/alphafold2/current/2.3/bfd/bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt \
--uniclust30_database_path=/shared/bank/alphafold2/2022-09-21/uniclust30/uniclust30_2018_08/uniclust30_2018_08 \
${param}
Thanks in advance and have a nice day.
Hi ,
That normal it seem that you are using alphafold/2.2.3 but calling the parameters for the v2.3, so load the v2.3.2 or change the path for the v2 params "/shared/bank/alphafold2/2022-09-21/params/" like for uniclust.
Hope that help.
Antoni
2 « J'aime »
Thank you so much for pointing it out, I was so absorbed changing the database paths that this escaped me!
However, with AlphaFold 2.3.2, I now encounter the same access right issues to the uniref30 databases as you...
I gave up trying to run AlphaFold 2.3.2 for now and am using v 2.2.3. The script below seems to work with the multimer mode.
module load alphafold/2.2.3
mkdir -p /tmp/$USER_alphafold
if [[ "${mode}" == "monomer" ]]; then
param='--pdb70_database_path=/shared/bank/alphafold2/2022-09-21/pdb70/pdb70'
elif [[ "${mode}" == "multimer" ]]; then
param='--uniprot_database_path=/shared/bank/alphafold2/2022-09-21/uniprot/uniprot.fasta --pdb_seqres_database_path=/shared/bank/alphafold2/2022-09-21/pdb_seqres/pdb_seqres.txt'
else
param='--pdb70_database_path=/shared/bank/alphafold2/2022-09-21/pdb70/pdb70'
fi
srun run_alphafold.sh --fasta_paths="${path}/${query_file}" \
--output_dir="${path}/${out_dir}" \
--model_preset="${mode}" \
--use_gpu_relax=True \
--db_preset=full_dbs \
--data_dir=/shared/bank/alphafold2/2022-09-21/ \
--uniref90_database_path=/shared/bank/alphafold2/2022-09-21//uniref90/uniref90.fasta \
--mgnify_database_path=/shared/bank/alphafold2/2022-09-21/mgnify/mgy_clusters_2018_12.fa \
--template_mmcif_dir=/shared/bank/alphafold2/2022-09-21/pdb_mmcif/mmcif_files \
--max_template_date=2020-05-14 \
--use_precomputed_msas=True \
--obsolete_pdbs_path=/shared/bank/alphafold2/2022-09-21/pdb_mmcif/obsolete.dat \
--bfd_database_path=/shared/bank/alphafold2/2022-09-21/bfd/bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt \
--uniclust30_database_path=/shared/bank/alphafold2/2022-09-21/uniclust30/uniclust30_2018_08/uniclust30_2018_08 \
${param}
Thanks again.
Hello,
If you don't mind can you please share the script for Monomar mode, I have been trying but it doesn't work.
Thanks in advance
Have a nice day!
Here is my script
#!/bin/bash
#SBATCH --partition=gpu
#SBATCH --gres=gpu:3g.20gb:1
#SBATCH --cpus-per-task=10
#SBATCH --mem=50G
#SBATCH --account=ancest_or_
#SBATCH --time 36:00:00
module load alphafold/2.2.3
mkdir -p /tmp/$USER_alphafold
srun run_alphafold.sh --use_gpu_relax=True --fasta_paths=/shared/projects/ancest_or_/SfruGR61.fa,SfruGR62.fa,SfruGR100.fa,SfruGR126.fa
--output_dir=/shared/projects/ancest_or_
--model_preset=monomer
--db_preset=full_dbs
--data_dir=/shared/bank/alphafold2/current/2.3
--uniref90_database_path=/shared/bank/alphafold2/current/2.3/uniref90/uniref90.fasta
--mgnify_database_path=/shared/bank/alphafold2/current/2.3/mgnify/mgy_clusters_2018_12.fa
--pdb70_database_path=/shared/bank/alphafold2/current/2.3/pdb70/pdb70
--template_mmcif_dir=/shared/bank/alphafold2/current/2.3/pdb_mmcif/mmcif_files
--max_template_date=2020-05-14
--obsolete_pdbs_path=/shared/bank/alphafold2/current/2.3/pdb_mmcif/obsolete.dat
--bfd_database_path=/shared/bank/alphafold2/current/2.3/bfd/bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt
--uniclust30_database_path=/shared/bank/alphafold2/2022-09-21/uniclust30/uniclust30_2018_08/uniclust30_2018_08
Hi,
For monomer I used alphafold 2.3.2 with the same db you used, excepted of uniclust who became uniref30, and to override the permission issues I've used the old dB and this seems to work :
--uniref30_database_path=/shared/bank/alphafold2/2022-09-21/uniclust30/uniclust30_2018_08/uniclust30_2018_08 \
What error is return with your script? That might help us to provide better answers.
Antoni
Thank you, I made the changes as yoyouentioned but after sometime it got killed. I have also attched the Error message both from my old and New script. Please, kindly let me know what changes should I make in my script?
Thanking you
Here is my Script (After correction)
#!/bin/bash
#SBATCH --partition=gpu
#SBATCH --gres=gpu:3g.20gb:1
#SBATCH --cpus-per-task=10
#SBATCH --mem=50G
#SBATCH --account=ancest_or_
#SBATCH --time 36:00:00
module load alphafold/2.3.2
mkdir -p /tmp/$USER_alphafold
srun run_alphafold.sh --use_gpu_relax=True --fasta_paths=/shared/projects/ancest_or_/SfruGR279.fa,SfruGR15.fa,OfurGR47.fa,OfurOR34.fa
--output_dir=/shared/projects/ancest_or_
--model_preset=monomer
--db_preset=full_dbs
--data_dir=/shared/bank/alphafold2/current/2.3
--uniref30_database_path=/shared/ bank/alphafold2/2022-09-21/uniclust30/uniclust30_2018_08/uniclust30_2018_08
--mgnify_database_path=/shared/bank/alphafold2/current/2.3/mgnify/mgy_clusters_2018_12.fa
--pdb70_database_path=/shared/bank/alphafold2/current/2.3/pdb70/pdb70
--template_mmcif_dir=/shared/bank/alphafold2/current/2.3/pdb_mmcif/mmcif_files
--max_template_date=2020-05-14
--obsolete_pdbs_path=/shared/bank/alphafold2/current/2.3/pdb_mmcif/obsolete.dat
--bfd_database_path=/shared/bank/alphafold2/current/2.3/bfd/bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt
--uniclust30_database_path=/shared/bank/alphafold2/2022-09-21/uniclust30/uniclust30_2018_08/uniclust30_2018_08
Hi,
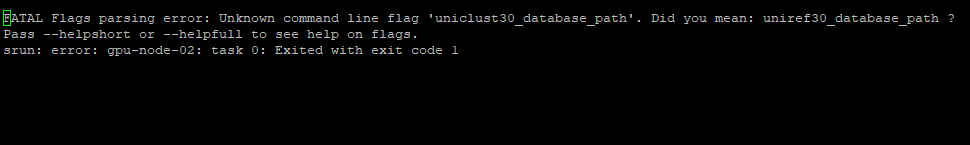
I think you have to delete the "--uniclust30_database_path" flag from your script, it is not compatible with Alphafold v2.3.2.
In case you are still interested by my script, it can run either on monomer or polymer mode, you can pick it on the command line. But it runs Alphafold 2.2.3 instead, so Antoni's suggestion might be better for you.
Have a nice day.
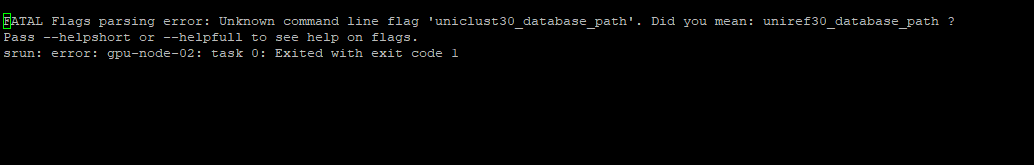
I got a error even after correction. I have attached the error screenshot for reference. Please could you have a look, I really appreciate it.
Thanking you
Here is my script (After correction)
#!/bin/bash
#SBATCH --partition=gpu
#SBATCH --gres=gpu:3g.20gb:1
#SBATCH --cpus-per-task=10
#SBATCH --mem=50G
#SBATCH --account=ancest_or_
#SBATCH --time 36:00:00
module load alphafold/2.3.2
mkdir -p /tmp/$USER_alphafold
srun run_alphafold.sh --use_gpu_relax=True --fasta_paths=/shared/projects/ancest_or_/SfruGR61.fa,SfruGR62.fa,SfruGR100.fa,SfruGR126.fa
--output_dir=/shared/projects/ancest_or_
--model_preset=monomer
--db_preset=full_dbs
--data_dir=/shared/bank/alphafold2/current/2.3
--uniref30_database_path=/shared/bank/alphafold2/2022-09-21/uniclust30/uniclust30_2018_08/uniclust30_2018_08
--mgnify_database_path=/shared/bank/alphafold2/current/2.3/mgnify/mgy_clusters_2018_12.fa
--pdb70_database_path=/shared/bank/alphafold2/current/2.3/pdb70/pdb70
--template_mmcif_dir=/shared/bank/alphafold2/current/2.3/pdb_mmcif/mmcif_files
--max_template_date=2020-05-14
--obsolete_pdbs_path=/shared/bank/alphafold2/current/2.3/pdb_mmcif/obsolete.dat
--bfd_database_path=/shared/bank/alphafold2/current/2.3/bfd/bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt
--uniclust30_database_path=/shared/bank/alphafold2/2022-09-21/uniclust30/uniclust30_2018_08/uniclust30_2018_08 \
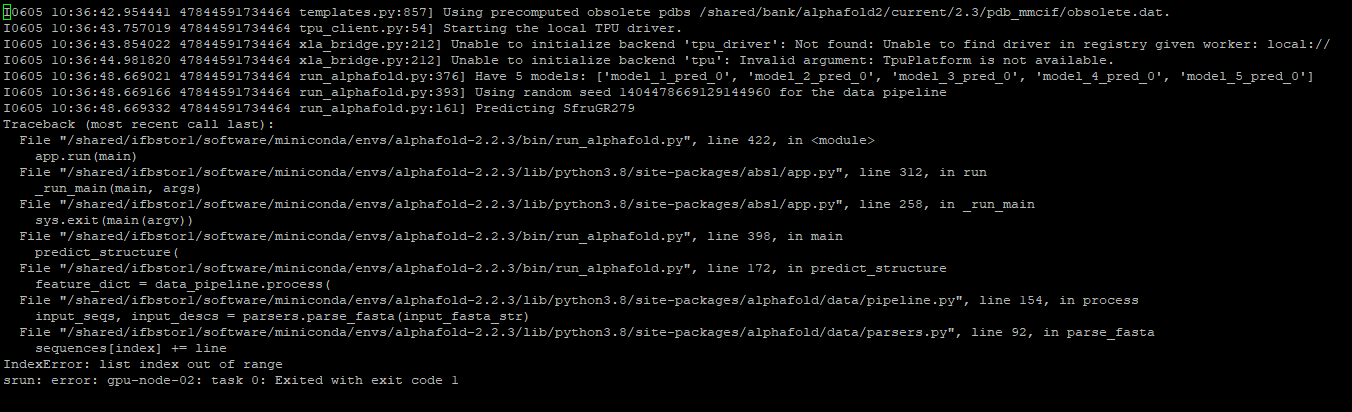
I also got a error from running Alphafold 2.2.3. I have attached the error screenshot for reference. Please could you have a look, I really appreciate it.
Thanking you
Here is my script (After correction)
#!/bin/bash
#SBATCH --partition=gpu
#SBATCH --gres=gpu:3g.20gb:1
#SBATCH --cpus-per-task=10
#SBATCH --mem=50G
#SBATCH --account=ancest_or_
#SBATCH --time 36:00:00
module load alphafold/2.2.3
mkdir -p /tmp/$USER_alphafold
srun run_alphafold.sh --use_gpu_relax=True --fasta_paths=/shared/projects/ancest_or_/SfruGR279.fa,SfruGR15.fa,OfurGR47.fa,OfurOR34.fa
--output_dir=/shared/projects/ancest_or_
--model_preset=monomer
--db_preset=full_dbs
--data_dir=/shared/bank/alphafold2/current/2.3
--uniref90_database_path=/shared/bank/alphafold2/current/2.3/uniref90/uniref90.fasta
--mgnify_database_path=/shared/bank/alphafold2/current/2.3/mgnify/mgy_clusters_2018_12.fa
--pdb70_database_path=/shared/bank/alphafold2/current/2.3/pdb70/pdb70
--template_mmcif_dir=/shared/bank/alphafold2/current/2.3/pdb_mmcif/mmcif_files
--max_template_date=2020-05-14
--obsolete_pdbs_path=/shared/bank/alphafold2/current/2.3/pdb_mmcif/obsolete.dat
--bfd_database_path=/shared/bank/alphafold2/current/2.3/bfd/bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt
--uniclust30_database_path=/shared/bank/alphafold2/2022-09-21/uniclust30/uniclust30_2018_08/uniclust30_2018_08 \
Hi,
just to infrom that now my alphafold is wroking. Thank you evryone for the help