Good morning,
I have some issues with Jupyter hub that I was trying to solve in these days without any success.
ISSUE 1
When I am trying to run a jupyter notebook, this error appears
Error Starting Kernel
Unhandled error
ISSUE 2
I cannot save Jupyter notebook
File Save Error for EQA_report.ipynbUnexpected error while saving file: shared/projects/emergen_hera_eqa/quality-assessment-hera.backup/src/EQA_report.ipynb unable to open database file
ISSUE 3
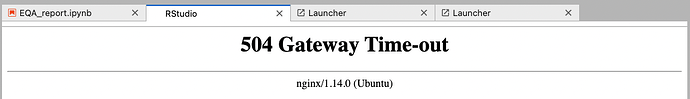
I cannot use Rstudio
How can I solve this errors?
If it is useful, I am available for a visio.
Merci beaucoup,
Arianna Tonazzolli
Dear Arianna,
You have reached the maximum number of files on your home directory.
RStudio/Jupyter have to create some files on your home directory and so failed to start.
There are two kinds of quota:
- disk quota: limit amount of data you can store.
- number of files quota: limit number of files you can create (on the IFB core cluster, this limit is 1000 files per GB)
You have 150000 files on /shared/home/atonazzolli:
$ lfs quota -h -p 162809 /shared/home/atonazzolli
Disk quotas for prj 162809 (pid 162809):
Filesystem used quota limit grace files quota limit grace
/shared/home/atonazzolli
6.179G 100G 150G - 150000* 100000 150000 -
So you need to clean this directory. It's often due to conda environnement (which create a lot of files).
Or you can move files on your project directories.
Best regards
Dear dbenaben,
Thanks for your quick reply.
I have already removed some files/folders before to create more space. I am worry to remove important files for the server. Can I remove .conda folder or it could generate problems?
Merci,
Arianna
You can remove your .conda directory but all your conda installations (packages and environments) will be removed.
If you know which packages and environments you have created, you can easily recreate it.
But before delete the directory, you can just try to list and remove some unused environment:
$ conda env list
$ conda env remove <env>
I looked a little bit more... and in your case, you have also lot of local installation files:
#Files Directory
72577 /shared/home/atonazzolli/.local
40453 /shared/home/atonazzolli/.planemo
# And more specifically:
65542 /shared/home/atonazzolli/.local/lib/python3.9
40369 /shared/home/atonazzolli/.planemo/gx_venv_3
Maybe you can have a look on this....
Thank you so much for your help.
I have removed some files and ISSUE 1 and 2 are solved. However, I am still not able to open R studio. Do I need to remove more files?
Merci,
Arianna
You have yet 108506 files (quota is 100k).
RStudio still return "error (Disk quota exceeded)" for your account.
Could you clean/move more files and retry RStudio ?
It still not work.
lfs quota -h -p 162809 /shared/home/atonazzolli
Disk quotas for prj 162809 (pid 162809):
Filesystem used quota limit grace files quota limit grace
/shared/home/atonazzolli
4.072G 100G 150G - 62773 100000 150000 -
Sorry again but unfortunately a new error appears now
File Save Error for EQA_report.ipynb
Unexpected error while saving file: shared/projects/emergen_hera_eqa/quality-assessment-hera.backup/src/EQA_report.ipynb attempt to write a readonly database
Thanks,
Arianna
I don't understand this error.
Could you try to relaunch a Jupyter Job (Hub Control Panel > Stop > Start):
And:
And finally relaunch a job.
When you say that RStudio is not working, do you try inside JupyterHUB or with https://rstudio.cluster.france-bioinformatique.fr/ ?
I followed your instructions and now everything is working, thanks a lot 
Hi @dbenaben
Thanks a lot for your help, this apparently solved Arianna's problem. I realised that my recurrent problems with the RStudio server might result from the same cause.
I would like to use the same command as Arianna in order to check my quota, but I did not understand which PID I should use.
I attempted to use the same as her:
(base) [jvanhelden@clust-slurm-client ~]$ lfs quota -h -p 162809 /shared/home/jvanhelden
Disk quotas for prj 162809 (pid 162809):
Filesystem used quota limit grace files quota limit grace
/shared/home/jvanhelden
4.489G 100G 150G - 106815* 100000 150000 6d4h34m8s
I obtain 106815, which exceeds the quota. However, when I re-run the command after having suppressed several thousands of files I still have the same number of files (106815).
Can you please indicate me how to choose the right PID ?
Thanks a lot
Jacques
Hello Jacques,
Indeed, it's a bit tricky to request the quota (for your home or project) with Lustre (file system used on the IFB storage). You need to know the project ID (-p) associated with the directory...
So, Gildas developed status_bars to have, among other things, some space storage information (but not the number of files).
And we use a bash function (thanks Gildas again) to have it quickly (we should add this kind of command for users).
lfsgetquota bash function
# Need some sudo permission
lfsgetquota () {
gid=$(getent group $1 | cut -f 3 -d ':')
echo "GID: $gid"
if [ -d /shared/projects/$1 ]
then
echo "lfs quota -h -p $gid /shared/projects/$1"
sudo lfs quota -h -p $gid /shared/projects/$1
fi
if [ -d /shared/home/$1 ]
then
echo "lfs quota -h -p $gid /shared/home/$1"
sudo lfs quota -h -p $gid /shared/home/$1
fi
}
In your case, for your home directory, the project ID is 100073.
So you have to use lfs quota -h -p 100073 /shared/home/jvanhelden.
Thanks a lot, David
Apparently I use a lot of space (there is one big project I should move) but not too many files.
(base) [jvanhelden@clust-slurm-client src]$ lfs quota -h -p 100073 /shared/home/jvanhelden
Disk quotas for prj 100073 (pid 100073):
Filesystem used quota limit grace files quota limit grace
/shared/home/jvanhelden
1.429T 2T 2.1T - 22168 2000000 3000000 -
Yes exactly. So It's not a quota issue with RStudio.
Unfortunately, I don't understand what's going wrong (there is a "Permission denied" on a Rproj file on the emergen_hera_eqa project but I don't understand why).
I kill your session on RStudio server. Could you retry ?