This topic is dedicated to tips concerning the use of workflows in Galaxy that may help you while performing analyses with Workflow4Metabolomics. Do not hesitate to glance over it, in particular prior to any help request concerning workflow-related issues.
I imported a workflow from another Galaxy instance but when I wanted to run it or edit it in usegalaxy.fr, I got some error/warning messages.
This can happen when the needed tools or tool versions are not available in usegalaxy.fr. In particular, if your imported workflow contains old tools or versions, information about these tools may not be retrievable. Thus, you may obtain one of the following:
The tool exists however the version that have been used is not available - the tool box is displayed in your workflow but with default parameter values.
In this configuration, there are two main possibilities:
- You know what were the parameters you used (e.g. you saved the information somewhere, or the Galaxy instance you previously used is still available and you can check the concerned workflow on it). In this case, we encourage you to use the version currently available on usegalaxy.fr (if it is a newer version), manually checking the parameters that have been set to default and retesting it on a dataset you know. Indeed, in a workflow approach we highly recommand to try being up-to-date to ensure being at the best potential of science.
- There is no way for you to get access to the previous parameters AND/OR the version you used was a newer one compared to the one available on usegalaxy.fr AND/OR you tested the version installed on usegalaxy.fr as recommanded but it does not deliver the results you need anymore. In this case, we invite you to post an installation request on the forum. Please precise in your post (i) the concerned tool name, (ii) the associated version, (iii) which instance you imported your workflow from, (iv) why you need this version installed (e.g. not able to get the parameters, need of this specific version for result purposes...).
The tool is missing. You get ‘red boxes’ in your workflow.
This generally leads to two configurations:
- The tool you used in the other Galaxy instance is substancially old; a newer version of the tool is available but it is not automatically linked to the old one inside Galaxy. Please take some time to check whether this is the case searching through the tool panel. If it is confirmed, then follow the procedure previously described for versions that are not available, adding the tool in your workflow as if all parameters were put to default.
- The tool you used is not available in usegalaxy.fr, in any version. In this case, we invite you to post an installation request on the forum that the support team will examine. Please precise in your post (i) the concerned tool name, (ii) the associated version, (iii) which instance you imported your workflow from, (iv) a short description of what this tool is designed for and in which tool panel category it would be relevant.
Note: if you updated your imported workflow, do not forget to save it afterwards. You can even re-export it from usegalaxy.fr if you need to keep traceability for any reason.
I wanted to extract a workflow from one of my histories but I ran into an error message when clicking on the "Create Workflow" button, saying:
Internal Server Error
Galaxy was unable to successfully complete your request
An error occured.
This error message is generic and it may be difficult for you to determine why the issue occured. When dealing with workflow extraction from histories, one known issue could be the following:
- An internal Galaxy bug fractionned one of your job in distinct parts.
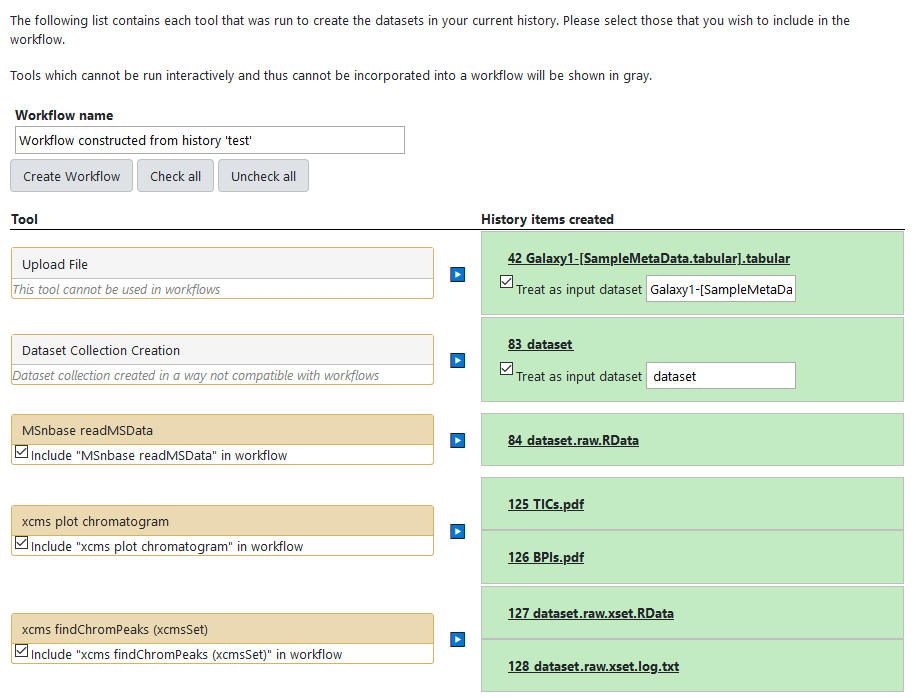
To check whether this has happenned in your case, have a close look at the central panel list. What you should have is one "left box" per input data or job, and one "right box" per generated dataset. Note that the generated dataset should be gathered by corresponding jobs as the following:
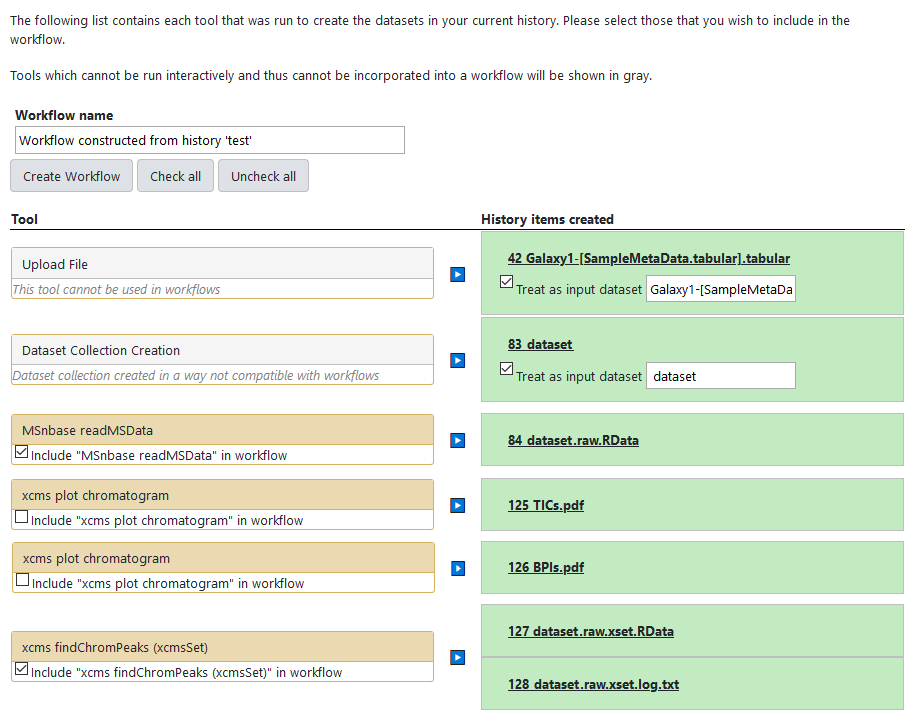
In some very rare cases, an internal error may have happened somehow, dividing one single job into two, as it would be in the example bellow where the "xcms plot chromatogram" job appears as two seperated jobs (one per input dataset):
If this happens, Galaxy can not extract the workflow as long as it includes the bugged job. The solution here is to untick the concerned step as bellow:
Once the workflow is extracted, you need to complete it again directly in the workflow "Edit" mode, by manually adding the concerned tool (putting the right parameters and links to other tools).
If you carefully looked at your jobs and do not notice any similar problem, do not hesitate to post a help request. Please precise what you already checked in your request description.