Dear workflow4metabolomics community,
since I got GCMS measurements of some plant leaf material I tried to get some untargeted analyis going using your platform with xcms and metaMS. I worked through some of Your tutorials but unfortunatelly I am not able to optimize my parameters within the workflow to a point where I dont get a lot of zeros within my list of unknowns I detect in my samples. I even tired the IPO package from bioconductor on our linux server to help me, but the outcome there was even worse.
Therefore You could make me a happy PhD student, if someone with more experience would give it a look and give me some feedback. I attach the workflow as well as a history with the raw files and the outcome of my workflow so far.
Workflow
Data
Thank You very much for Your help!
Hi,
Finding the "right" peakpicking parameter can be a real adventure depending on the nature of your data. Usually a dataMatrix with a lot of zero means that :
1- your samples are really differents from each others
2- that your similarity threshold is to high and cause metaMS to split peaks
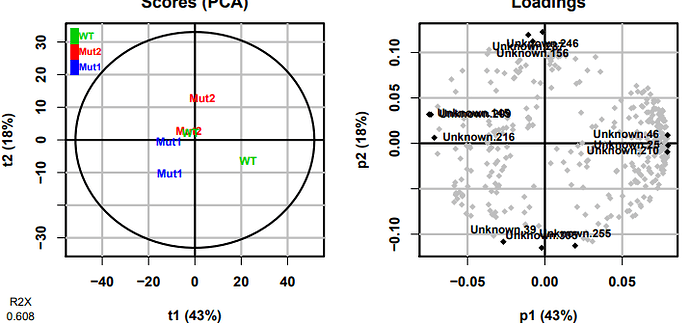
I've quickly looked at your data and for a first try not that bad , in fact you only have 0 for low intensity compounds and even here a would say not that much zero in fact
here is a PCA made in Galaxy showing that your close to something (I think)
for me, to go further in the dataprocessing we need more samples and also QC samples to be able to correct for intensity deviation.
Do you have such data to add to the process?
Yann
Dear Yann,
Thank You very much for the quick and helpful response 
Happy to hear, that I am on the right track.
I have more samples from different timepoints and we measure a few alkane standards every time the GCMS runs. Is that what you mean by QC-Samples? How do I correct for intensity deviation with it?
Additionally I notices, that my peaktable from MetaMS often shows "8388096" as a value for different unknowns, which does not make sense to me, maybe You have an idea.
Here is another timepoint, already processed in the workflow and the alkane standards we measure:
With appreciation
Malte