Hello,
I recently started pre-processing LCMS data and encountered a problem at the fillPeaks step ("This job was terminated because it ran longer than the maximum allowed job run time.").
I have a very large sample size (239 samples with a total of 63838 lines after grouping-retcor-regrouping) which might explain why the job exceeded the maximum allotted run time.
Does anyone have an idea of what could be the reason behind the error (if the problem comes from the settings or dataset) ? If the problem doesn't come from either of those, what could be done to make sure that the fillpeak step can proceed without any problems ?
The relevant job API ID is e97778feb92a1bde.
Thank you for any solution or advice,
L. Brasseur
Hello,
I guess the process is too time-consuming relatively to the job time allowed with usegalaxy.fr. I have no idea what value this maximum limit is set to, so I can not help for this part and maybe someone else from @team.w4m can answer that.
Nonetheless, with your dataset size, I am not surprised that fillChromPeaks could take quite a time to run. However, maybe this time could be reduced significantly if you consider revising your previous XCMS parameter choices (if relevant).
The thing is, the more "NA" you have in your dataset, the more time is needed for fillChromPeaks to run. With your high number of variables (63838), one can wonder whether these variables may be initially found only in a few samples, generating a lot of "NA" to fill. Depending on your design and research question, it could be important to have variables that only concerns a few samples, but it is quite common that you may only really need variables that are shared at least by some groups of samples in you design. There are options in XCMS that allow to filter, in the groupChromPeak step, peaks that are not found numerous enough in at least a group of samples. Maybe considering using these functionality could be a good way to reduce the number of variables (reducing the number of NA) with relevance.
Another thing could be to review whether some ions have been artificially splitted in several variables. This can increase the number of variables with lots of NA, while not corresponding to distinct signals. This can be checked with the results from groupChromPeaks, and if it turn out to be the case the best is to review the parameters used to try to have a more appropriate result, which may lead to less NA values thus a less time-consuming fillChromPeaks.
These are only hypotheses, not being aware of your project nor results. It is possible that you already have customed your previous XCMS steps with care, and in this case actions have to be taken from the usegalaxy part. However, if your run was a first attempt before begining the optimisation process of your XCMS extraction, I highly recommand that you first customise you previous XCMS steps, and only try the fillChromPeak module once the previous steps are settled to the final parameter choices.
Best regards,
Mélanie
Thank you for your answer.
I've tried reducing the amount of metabolites with the groupChromPeaks step and despite a 30% or so decrease of the amount of lines (from 64k to about 45k) the fillpeaks step seems to still take longer than the allotted time (24 hours). I suppose the large amount of data makes this step impossible via W4M with the current settings I'm using.
Best regards,
L. Brasseur
Thank you for your feedback. Do you have an idea of the proportion of NA in your table prior to the fillChromPeak step? If needed, you can use the Intensity_check Galaxy module to get some NA sumup indices per ion.
Concerning the runtime limit, @gildaslecorguille do you know if it is possible to have an extension of it?
Mélanie
I don't have a specific number in mind but most of what I have seems to be made of NAs (to the point where a CTRL+F "NA" command on excel just crashes the software entirely).
If Excel is a problem to have an idea, you can use the variableMetadata file to get the information in another way. Indeed, in this file you will find columns (one per class you consider in the extraction process, or a single one if no class was specified) that counts the number of samples in which each ion was found. Thus, with a simple trick of substraction (number of samples - content of the column), you can have the information of the number of NA per ion.
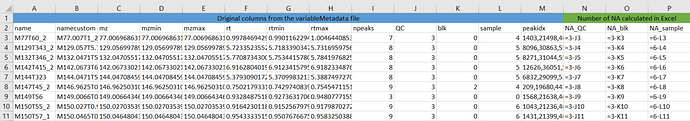
Here is an example:
Here I used 3 classes for extraction (QC, blk and sample) so I have 3 corresponding columns generated in the variableMetadata. I have respectively 3, 3 and 6 samples in these groups. In Excel, I get have the number of NA for each ion and for each group with the calculation given in columns N, O and P. The total number per ion is thus the sum of these 3 "green" columns.
This can be easier than the CTRL+F for huge dataMatrix in Excel. You can also add some metrics as the % of NA based on these columns. Please note that you can also use Galaxy to have this computed by using the Intensity_check Galaxy module, available in the "Quality processing all" section in the tool panel, with a graphical representation as complement.
Again, I do not know your project so my comment may not match your study design, but if you have that many NA in your data with good reason to keep all these ions in your dataset, it may be wise not to perform fillChromPeaks right away but first analyse your data with a "NA / not NA" point of view to have a better idea of what your are dealing with. Then, if your conclusion is that it is relevant to analyse all these ions in a more "quantitative" way, then consider trying to perform the fillChromPeaks. Trying to fill NA in a dataset where NA is the majority value is always risky and should be done with huge care.
Anyway, still waiting for a Galaxy guy to give us some light on what is possible regarding the runtime limit 
Mélanie
I was planning on filtering the NAs later on (present in 50% or even 75%) after this step and CAMERA. Though I doubt that CAMERA would run correctly if the fillpeaks step doesn't manage to finish processing in time.
The walltime by default is 24 hours.
We can increase it if you judge necessary!
I managed to fix the Fillpeaks part of the problem by drastically reducing the amount of lines I have but now CAMERA.annotate is the one posing a problem with exceeding run times. I'd greatly appreciate the time extension!
FillChromPeaks and CAMERA.annotate can long for 2 days instead of 1.